Back to the old days, I’ve studied how to implement highly efficient PyTorch pipelines for multi-gpu training [1]. DistributedDataParallel is the way to go, but it is cumbersome that we need boilerplates for spawning workers and constructing data readers.
Now, PyTorch Lighting offers clean API for setting up multi-gpu training easily. Here is a template I designed, which I will stick to for prototyping models for the rest of my life : )
import os
from typing import List, Any
from dataclasses import dataclass
import torch
import torch.distributed as dist
from torch import nn
from torch.nn import functional as F
from torch.utils.data import DataLoader, IterableDataset, random_split
from torchvision.datasets import MNIST
from torchvision import transforms
import pytorch_lightning as pl
from pytorch_lightning.metrics.functional import accuracy
TOTAL_NUM_BATCHES = 320
BATCH_SIZE = 32
STATE_DIM = 5
NUM_GPUS = 2
NUM_WORKERS = 2
UPDATE_FREQ = 1
WEIGHTS = torch.tensor([2.0, 3.1, 2.1, -1.5, -1.7])
@dataclass
class PolicyGradientInput:
pid: int
state: torch.Tensor
reward: torch.Tensor
def __len__(self):
return self.state.shape[0]
@classmethod
def from_batch(cls, x):
return cls(
pid=x[0][0].item(),
state=x[1][0].reshape(BATCH_SIZE, STATE_DIM),
reward=x[2][0].reshape(BATCH_SIZE, 1),
)
class EpisodicDataset(IterableDataset):
def __iter__(self):
worker_info = torch.utils.data.get_worker_info()
pid = os.getpid()
if worker_info is None:
total_num_batches = TOTAL_NUM_BATCHES // NUM_GPUS
worker_id = -1
else:
total_workers = worker_info.num_workers
total_num_batches = int(TOTAL_NUM_BATCHES // NUM_GPUS // total_workers)
worker_id = worker_info.id
# You will see that we have an EpisodicDataset on each of NUM_GPUS processes
print(f"{worker_info}, pid={pid}, total_num_batches={total_num_batches}")
for _ in range(total_num_batches):
state = torch.randn(BATCH_SIZE, STATE_DIM)
reward = torch.sum(state * WEIGHTS, dim=1)
yield (pid, state, reward)
class PPO(pl.LightningModule):
def __init__(self):
super().__init__()
self.model = nn.Sequential(
nn.Linear(STATE_DIM, 128), nn.ReLU(), nn.Linear(128, 128), nn.ReLU(), nn.Linear(128, 1)
)
self.traj_buffer = []
self.update_freq = UPDATE_FREQ
self.step = 0
def training_step(self, batch: List[Any], batch_idx):
batch: PolicyGradientInput = PolicyGradientInput.from_batch(batch)
self.traj_buffer.append(batch)
self.step += 1
rank = dist.get_rank()
# use first three trajectories' pids as as signature. quickly check if all trainers share the same data
# the answer is each trainer maintains different trajectories
traj_buffer_signature = ','.join([str(traj.pid) for traj in self.traj_buffer[:3]])
print(f"rank={rank}, traj_buffer_len={len(self.traj_buffer)}, step={self.step}, signature={traj_buffer_signature}")
if self.step % self.update_freq == 0:
model_params = list(self.model.parameters())[0][0].detach().cpu().numpy()
print(f"before {self.step} step training: rank={rank}, model_params={model_params}")
return self.update_model()
def configure_optimizers(self):
# Somehow, Adam doesn't work for linear regression
# optimizer = torch.optim.Adam(self.parameters(), lr=1e-2)
optimizer = torch.optim.SGD(self.parameters(), lr=1e-2)
return optimizer
def update_model(self):
traj = self.traj_buffer[-1]
loss = F.mse_loss(self.model(traj.state), traj.reward)
rank = dist.get_rank()
print(f"rank={rank}, step={self.step}, loss={loss}")
return loss
ds = EpisodicDataset()
dl = DataLoader(ds, batch_size=1, num_workers=NUM_WORKERS, pin_memory=True)
ppo = PPO()
trainer = pl.Trainer(gpus=NUM_GPUS, max_epochs=1, progress_bar_refresh_rate=1, accelerator='ddp')
trainer.fit(ppo, dl)
As you can see, you only need to define a Dataset, a DataLoader with appropriate NUM_WORKERS, and a pytorch-lightning Trainer in which you specify the number of gpus. Each of the NUM_GPUS GPUs will then use NUM_WORKERS processes for reading data and use one main process for training the model.
The example shows that each trainer on a GPU maintains a list of trajectories, which is not shared with the trainers on other GPUs. However, I believe model parameters are synced after every loss function computation because the underlying mechanism is still DistributedDataParallel.
References
[1] https://czxttkl.com/2020/10/03/analyze-distributeddataparallels-behavior/

 .
. 












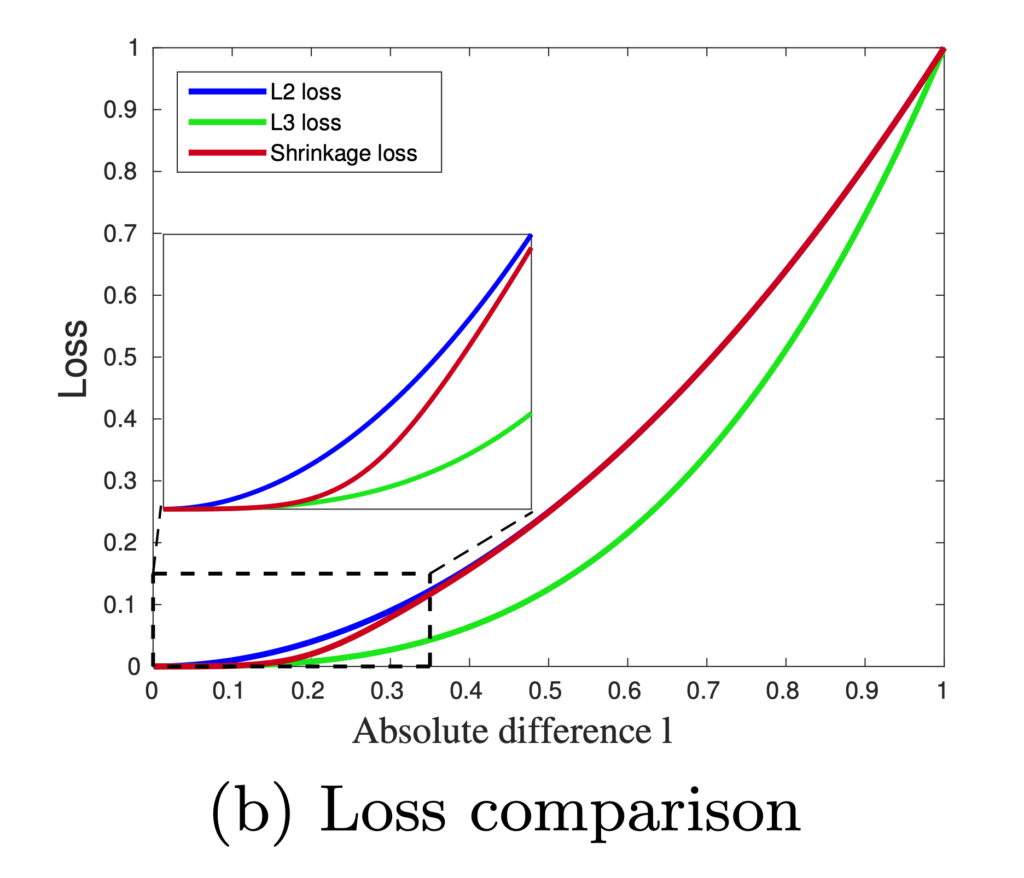
![Rendered by QuickLaTeX.com \[L_S = \frac{l^2}{1 + exp\left(a\cdot \left( c-l \right)\right)}\]](https://czxttkl.com/wp-content/ql-cache/quicklatex.com-60f04910d6082a4a6a347f51491eeac0_l3.png)